Synthesis of gene clusters, gene assemblies and multigenic constructs:
How the Design works:
It uses
powerful biocomputation to accomodate all the design requirements, some of which are default and others that are optional or customer-specific.

|
Basically, it is like ordering a la carte in a restaurant:
- pick your main course and side dishes from our menu,
- order your selection and
- let our chefs put together a palate pleaser for you.
- and enjoy your (multi-)gene assembly
|
The below graphic gives you an idea of what the design can look like. It can be designed to fully comply with the design parameters of the
Toggle plasmid system but it can
also be adjusted to any specific cloning / expression system you have, e.g. by considering specific restriction sites and eliminating duplicates computationally from the
overall design.
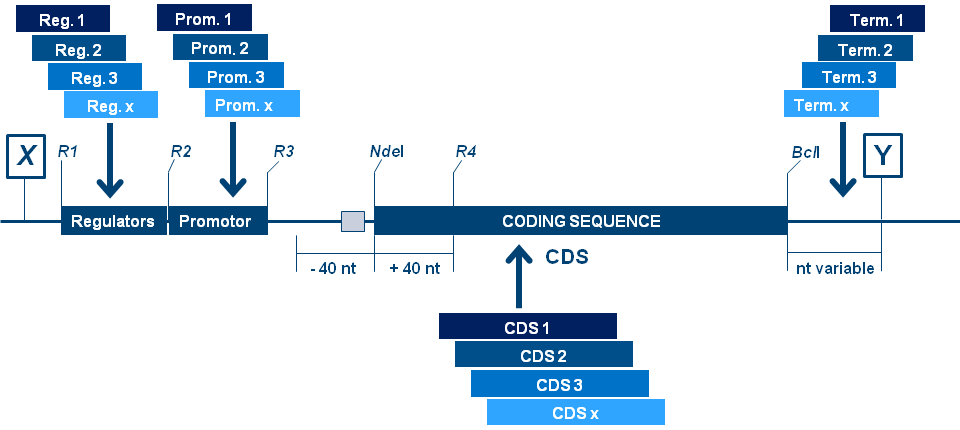
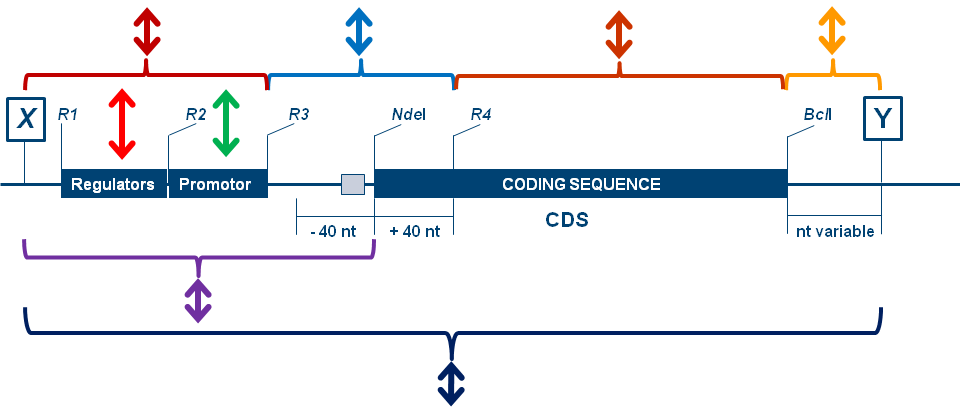
Figure 1.: Gene Cluster design strategy of DNA – assembling with de-assembling option and partial sequence substitution opportunity (ACDCS).
X and Y are AscI(BssHII) and MauBI(BssHII) respectively. Combined by ligation BssHII reconstitutes at the position of ligation.
This provides the opportunity for to generate multiple BssHII sites at the expression – box junction which further allows
you to de-assemble multi-genic constructs (gene clusters) which were generated this way.
R1..Rn are sites calculated out of all the coding sequences and also not present in the intergenic regions by design.
These are positioned only at desired locations in one of the individual gene constructs for the exchange of specific
parts of the intergenic regions (promoters, leaders, RBSs) and individual CDS or variants thereof. 3'-of X and 5'-of Y specific recombination
sites are serving for the option of recombination strategies for assembly of pathways.

The beauty of the design lies in the fact that by precisely defining and inserting unique restriction sites you can specifically exchange selected constructional elements,
e.g. promoters, terminators, enhancers, the leader or translation initiation region (+/- 40 nt around the start ATG). This way you can build your own
repository of
genetic building blocks.
You can find more information on design criteria and parameters in the
Cluster Design section.

Gene clusters in general
Gene clusters have, on one side, evolved naturally, but can also be designed, e.g. for the purposes of industrial biotechnology. Whether natural or artificial,
these assemblies of genes usually code for tightly interacting and functionally coupled enzymes, e.g. those involved in complex, multi-step biochemical processes
(catalysis, degradation, etc.). Examples are enzyme cascades involved in amino acid synthesis, the artimisinin biosynthetic pathway, or polyketide synthase complex.

Establishing such pathways in a heterologous host is challenging as it involves various design constraints that need to be considered, e.g.:
- Codon use has to be adapted to the expression host.
- Promoters and control elements have to be redesigned to match host requirements, e.g. when transferring a gene from yeast to E.coli
- Expression of the individual genes has to be balanced to improve yield of the entire pathway.
- Specific requirements of the expression host's matabolisms have to be taken into consideration. While certain genes may work in one host,
they may wreak havoc (metabolic toxicity) in another host if they are not modified or kept in check. In addition, it may be critically important to shut down
other pathways that siphon off educts or products and thus negatively impact yield.
This list can be continued but the literature is rife with various examples of how to tackle such obstacles and problems.
Smart bioinformatic analysis and
gene cluster design can help address these bottlenecks.

ACDC-SD:
is ATG's
assembly cloning - disassembly cloning - substitution design. It is a stringent design that applies a uniform set of constructive principle to
genes, gene cassettes and gene clusters. In effect, this means that all modular elements will be fully compatible with other equivalent modules (for the same organism
or vector class).